

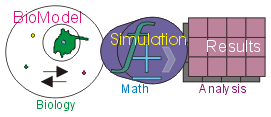
Virtual Cell Modeling and Simulation Framework at The National Resource for Cell Analysis and Modeling (NRCAM)
The National Resource for Cell Analysis and Modeling
(NRCAM),
developer of the Virtual Cell, is a national resource center supported
by the National Center for Research Resources (NCRR), at the National
Institutes
of Health (NIH).
Developed by the U.S. Department of Energy to complement its Genomes to Life project.
MCP takes a whole-genome approach to understanding the function and regulation of genes for a single living system and the pathways in which the protein products interact.
In the MCP, scientists will begin to write a
comprehensive
"owner's manual" for a microbial cell.Microbial cells have internal
organization
and complex control systems that allow them to respond to their
environment.
They can work as miniature chemistry laboratories, making unique
products
and carrying out specialized functions. Ultimately, understanding the
complex
functioning of a single microbial cell will enable science to go far
beyond
just exploiting the beneficial capabilities of microbes to meet DOE's
missions.
The knowledge gained will apply to cells in all living things. Thus the
MCP represents a first step in moving from cataloguing molecular parts
to constructing an integrative view of life at the level of a whole
organism
-- microbe, plant, or animal (Human Genome News vol 11, Nos 3-4, 2001).
The goal of
Project CyberCell is to create an accurate dynamic model of a simple
living
organism within 5 years
using a molecular-level population dynamics approach.
The overarching premise of Project CyberCell is
the integration of experiment and
theory, where directed, high-resolution biological and biomolecular
measurements
are used to drive and validate combinatorial numerical analysis
and systems modeling.
A computer software environment for modelling and simulation of a hypothetical cell.
Simulations can be run to conduct experiments in silico
Users can define:
Protein functions
Protein - protein interactions
Protein -DNA interactions
Regulation of Gene Expression
Cellular Metabolism Pathways
Designed to help predict consequences of changes in cell or environment, e.g.
- gene knockouts
- alteration of available metabolites
Consequences of intervention include:
- cell death
- changes in growth rate
- increase or decrease in expression of specific genes
In silico cell models complement experimental efforts to modify & engineer whole genomes
Enable modelling & simulation of the complex interactions among the gene products of whole genomes
E-cell model cosists of 3 lists:
- substance list: defines all objects making the cell and culture medium (substrates, products, catalysts)
- rule list: defines all the reactions that can take place within the cell
- system list: defines spatial spatial and/or functional structure of the cell and its environment
State of cell at each time frame expressed as:
- concentrations of all substances in cell
- cell volume, pH & temperature
In a single time interval each rule in the rule list is called upon to compute the change in concentration of each substance (proteins, DNA, RNA & small molecules) to generate next state of the cell.
Applications:
1. Assessment of the metabolic requirements of the cell
M. Genitalium currrently grown on complex material containing fetal bovine serum, yeast extracts
E-cell may help formulate chemically-defined synthetic medium
Comparison of experimental and computed results could result in identification of new enzmes or transporters
2. Decifering gene regulatory networks
Specific mechanisms for control of transcript levels may be identified by comparison of parallel in vitro and in silico experiments.
3. Definition of the minimum set of genes required under specific set of conditions
Non-essential genes can be modified by gene disruption
Features not modelled:
Proliferation, cell structure proteins
Reference: Tomita M et al. Bioinformatics
(1999) 15, 72-84
DIGITAL SIMULATION OF CELLULAR NETWORKS
Diagrammatic Cell LanguageTM
Complete diagrammatic language for large-scale network visualization and simulation
Colon cancer and other examples at Gene Network Sciences website
Physiolab and In Silico R&D (Entelos Corporation):
Provides a framework for integrating data (including
genomic,
proteomic, physiologic, and environmental) in the
context of a disease, with a focus on understanding
anddetermining
clinical responses to potential treatment.
DIGITAL SIMULATION OF EXPERIMENTS
Virtual experiments such as mutations and gene knock-outs.
Example: Digital
Disease ModelsTM
MATHEMATICAL MODELLING OF METABOLIC NETWORKS
Application of Flux Balance Analysis
1. Analysis of variation of the H. Influenzae Rd metabolic genotype in silico:
Schilling CH and Palsson BO (2000) J. Theor. Biol. 203, 249-283
Edwards JS and Palsson BO (1999) J. Biol.Chem. 274, 17410 -17416
Defined extreme pathways: systemically-independent biochemical pathways
Six different optimal metabolic phenotypes sould be defined
Redundant functions under defined conditions could be identified
2. Definition of system capabilities and capabilities of metabolism in E.Coli MG1655
Edwards JD and Palsson BO PNAS (2000) 97, 5528 - 5533
Applications:
1. Reconcile and curate sequence annotation by identifying unsupported reactions
2. Decifering gene regulatory networks
Specific mechanisms for control of transcript levels may be identified by comparison of parallel in vitro and in silico experiments. Coregulation and co-expression may be discovered
3. Definition of the minimum set of genes required under specific set of conditions
4. Identification of genes that can be deleted under critical conditions in specific human environments
Review:
Papin JA et al. (2003) Metabolic Pathways in the Post-genome era. Trends. Biochem. Sci. 28: 250-8
Lectures in Harvard Biophysics 101 Lecture Course:
Metabolic network flux models: Introduction to the basic concepts and linear optimization
Metabolic network flux models: Scientific and practical use
[Course
Schedule] [Metabolic
Pathways]
Bioinformatics
Programming
The Scriptome : a Minimal learning toolbox for Manipulating
Biological Data