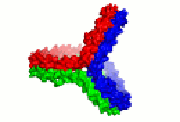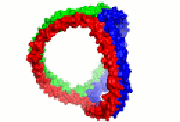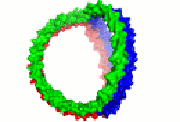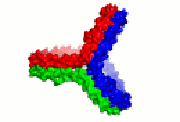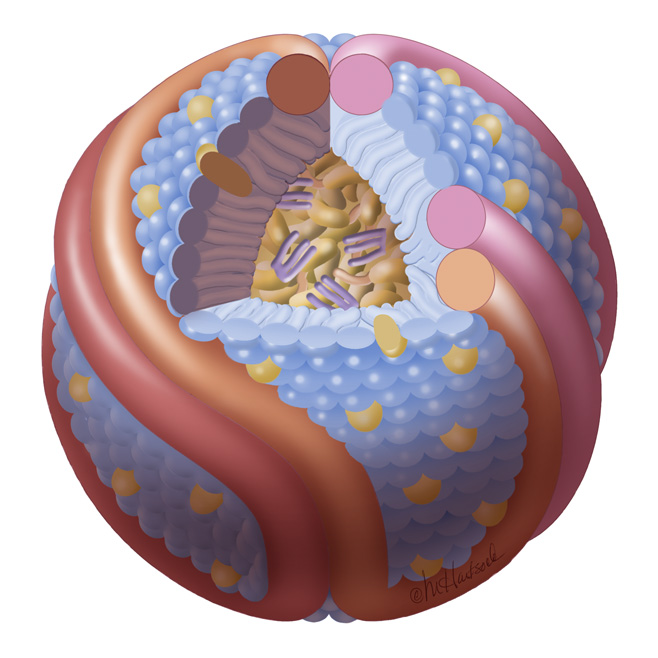
HDL cartoon |
Name:
HDL cartoon.
Description: We think this is a better representation of HDL composition and structure than most cartoons you will find on the internet. It is drawn approximately to scale with protein structures based on our work. Reference: Huang et al, Nature Structural and Molecular Biology 2011 4; 416-422. Image file (JPG, 658x658 pixels): Click here |

ApoA-I, classic 5/5 double belt. |
Name:
Classic 5/5 double belt model as described by Segrest
Description: Original double belt of two apoA-I molecules on a 96 angstrom rHDL particle. Helix 5 of each antiparellel belt overlap. Note: This model does not include residues 1-43. Reference: Segrest et al., J Biol Chem. 1999 Nov 5;274(45):31755-8. PDB file: Click here |
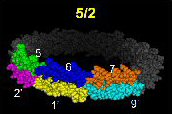
ApoA-I, alternative 5/2 double belt. |
Name:
Altered registry of 5/5 double belt model as proposed by us.
Description: Two apoA-I molecules on a 96 angstrom rHDL particle. Instead of helix 5 of each antiparellel belt overlapping, helix 5 overlapes with helix 2. Note: This model does not include residues 1-43. Reference: Silva et al, Biochemistry 2005 44(24); 8600-8607. PDB file: Click here |
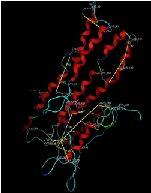
ApoA-I, lipid-free homology and cross-linking model. |
Name:
ApoA-I, lipid-free homology and cross-linking model.
Description: Full length apoA-I monomer folded as dictated by homology modeling and cross-linking with BS3. This was the first molecular detailed model of lipid-free apoA-I. Reference: Silva et al, Biochemistry 2005 44(8); 2759-2769. PDB file: Click here |
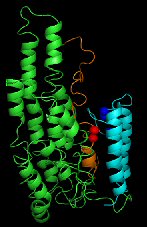
ApoA-IV, lipid-free homology and cross-linking model. |
Name:
ApoA-IV, lipid-free homology and cross-linking model.
Description: Full length apoA-IV monomer folded as dictated by homology modeling and cross-linking with BS3. This was the first molecular detailed model of lipid-free apoA-IV. Reference: Tubb et al, JBC 2008 283(25); 17314-17323. PDB file: Click here |
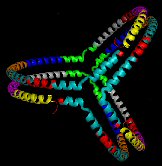
ApoA-I Trefoil in reconstituted spherical HDL. |
Name:
ApoA-I Trefoil.
Description: Three apoA-I molecules on a spherical HDL particle. Note: This model does not include residues 1-43 and may include breaks in the sequence. Reference: Silva et al, PNAS 2008 105(34); 12176-12181. PDB file: Click here |

ApoA-IV 64-335 dimer. |
Name:
ApoA-IV 64-335 dimer crystal structure 2.4 Angstroms.
Description: ApoA-IV 64-335 was crystallized as a dimer, revealing an interesting helix swapping dimeric interface. Reference: Deng et al, Structure 2012 20(5); 767-779. PDB file: Click here |
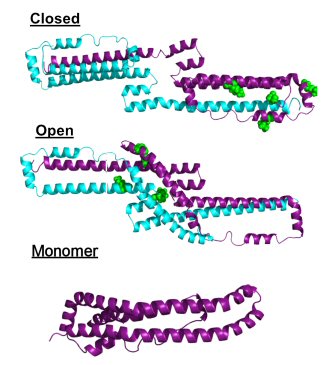
ApoA-I 1-185 dimers and monomer. |
Name:
ApoA-I 1-185 deletion mutant dimer and monomer.
Description: The deletion mutant of apoA-I that was crystallized by Mei et al was evaluated in solution both as a dimer and monomer by isotope-assisted chemical crosslinking and SAXS. Reference: Melchior et al, JBC 2016 291(10); 5439-5451. PDB file for "closed": Click here PDB file for "open": Click here PDB file for monomer: Click here |
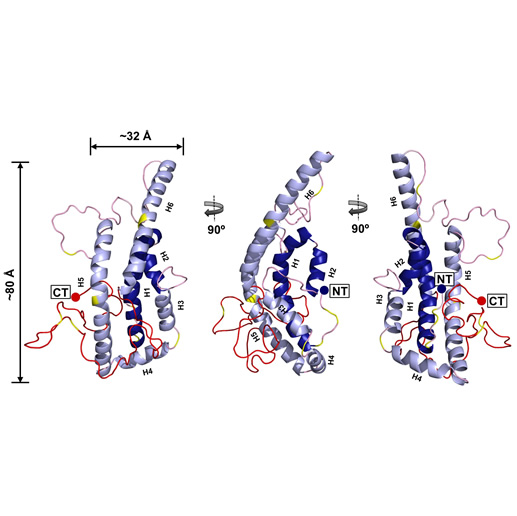
ApoA-I FL monomer. |
Name:
Full-length, lipid-free human apoA-I concensus model.
Description: This model was generated by combining cross-linking data from 3 laboratories as well as SAXS, HDX and X-ray crystallography data. It was generated in a collaborative effort from 8 different lipoprotein structural biology groups. Reference: Melchior et al, Nature Structure and Molecular Biology 2017 24:1093-1099 . PDB file: Click here |